|
|
|
| P R I N T E R - F R I E N D L Y F O R M A T | Return to Article |
D-Lib Magazine
November/December 2015
Volume 21, Number 11/12
Structured Affiliations Extraction from Scientific Literature
Dominika Tkaczyk, Bartosz Tarnawski and Łukasz Bolikowski
Interdisciplinary Centre for Mathematical and Computational Modelling, University of Warsaw, Poland
{d.tkaczyk@icm.edu.pl, bartek.tarnawski@gmail.com, l.bolikowski@icm.edu.pl}
DOI: 10.1045/november2015-tkaczyk
Abstract
CERMINE is a comprehensive open source system for extracting structured metadata from scientific articles in a born-digital form. Among other information, CERMINE is able to extract authors and affiliations of a given publication, establish relations between them and present extracted metadata in a structured, machine-readable form. Affiliations extraction is based on a modular workflow and utilizes supervised machine learning as well as heuristic-based techniques. According to the evaluation we performed, the algorithm achieved good results both in affiliations extraction (84.3% F1) and affiliations parsing (92.1% accuracy) tasks. In this paper we outline the overall affiliations extraction work flow and provide details about individual steps' implementations. We also compare our approach to similar solutions, thoroughly describe the evaluation methodology and report its results. The CERMINE system, including the entire affiliations extraction and parsing functionality, is available under an open-source licence.
Keywords: Metadata Extraction, Scientific Literature Analysis, Content Classification, Affiliations Extraction, Affiliations Parsing
1 Introduction
Academic literature is a very important communication channel in the scientific world. Keeping track of the latest scientific findings and achievements, usually published in journals or conference proceedings, is a crucial aspect of typical research work. Ignoring this can result in deficiencies in the knowledge related to latest discoveries and trends, which in turn can lower the quality of the research, make results assessment much harder and significantly limit the possibility of finding new interesting research challenges. Unfortunately, studying scientific literature, and in particular being up-to-date with the latest positions, is difficult and extremely time-consuming. The main reason for this is the very large and constantly growing volume of scientific resources, as well as the fact that publications are predominantly made available as unstructured text.
Modern digital libraries support the process of studying the literature by storing and presenting document collections not only in the form of raw sources of the documents, but accompanied with information related to citations, authors, organizations, research centres, projects, datasets, and also relations of various nature. Machine-readable data describing scholarly communication allows to build useful tools for intelligent search, detecting similar and related documents and authors, the assessment of the achievements of individual authors and entire organizations, identifying people and teams with a given research profile, and many more tools. In order to provide such high quality services a digital library requires access to the rich set of stored documents' metadata, including information such as authors and their institutions or countries. Unfortunately in practice, good quality metadata is not always available, is missing or fragmentary sometimes, or full of errors. In such cases the library needs an automatic method of extracting the metadata from documents at hand.
In this paper we propose a reliable method for extracting structured affiliations from scientific articles in PDF format. The algorithm is able to perform the following tasks:
- extracting authors and affiliations directly from a PDF article,
- associating authors with affiliations in the context of the article,
- detecting organizations, addresses and countries in affiliation strings.
The method is based on a modular work flow, which makes it easy to improve or replace one task implementation without changing other parts of the solution. We make extensive use of supervised machine-learning techniques, which increases the maintainability of the solution, as well as its ability to adapt to new document layouts.
Affiliation extraction and parsing functionality described in the article is part of CERMINE [14] — our open-source tool for automatic metadata and bibliography extraction from born-digital scientific literature. CERMINE web service, as well as the source code, can be accessed online [12].
In the following sections we describe the state of the art (Section 2), provide the details about the overall work flow architecture and individual implementations (Section 3) and finally report the evaluation methodology and its results (Section 4).
2 State of the Art
Even limited to analysing scientific literature only, the problem of extracting a document's authors and affiliations remains difficult and challenging, mainly due to the vast diversity of possible layouts and styles used in articles. In different documents the same type of information can be displayed in different places using a variety of formatting styles and fonts. For instance, a random subset of 125,000 documents from PubMed Central contains publications from nearly 500 different publishers, many of which use original layouts and styles in their articles. What is more, the PDF format, which is the most popular for storing source documents, does not preserve the information related to the document's structure, such as words and paragraphs, lists and enumerations, or the reading order of the text. This information has to be reverse engineered based on the text content and the way the text is displayed in the source file.
Nevertheless, there exist a number of methods and tools for extracting metadata from scientific literature. They differ in availability, licenses, the scope of extracted information, algorithms and approaches used, input formats and performance.
Han et al. [5] describe a machine learning-based approach for extracting metadata, including authors and affiliations, from the headers of scientific papers. The proposed method is designed for analysing textual documents and employs a two-stage classification of text lines with the use of Support Vector Machines and text-related features.
PDFX [2] is a rule-based system for converting scholarly articles in PDF format to XML representation by annotating fragments of the input documents. PDFX extracts basic metadata and structured full text, but unfortunately omits affiliation-related information. What is more, the system is closed source and available only as a web service.
GROBID [8] is a machine learning-based library focusing on analysing scientific texts in PDF format. GROBID uses Conditional Random Fields in order to extract a document's metadata (including parsed affiliations), full text and parsed bibliographic references. The library is open source.
SectLabel, a module from the ParsCit project [9, 3], also uses Conditional Random Fields to extract the logical structure of scientific articles, including the document's metadata, structured full text and parsed bibliography. The tool is able to extract affiliations, but does not parse them. ParsCit analyses documents in text format, and therefore does not use geometric hints present in the PDF files. The tool is available as open source.
Extracting and parsing affiliations is also provided by the NEMO system [6]. NEMO does not process PDF files directly, but rather extracts information from XML. What is more, affiliation parsing is done using regular expressions and dictionaries, which limits the possibility of adapting the solution to new formats and styles.
Enlil [4] is another tool able to extract authors, affiliations and relations between them from scientific publications. The system uses SectLabel [9] to detect authors and affiliations in the article's text, while splitting authors and affiliations lists, and associating authors with affiliations, are done by CRF and SVM classifiers. Unfortunately, Enlil does not perform affiliation parsing.
In terms of functionality, input format and methods used, our solution is most similar to GROBID. CERMINE extracts affiliations directly from PDF files, assigns them to extracted authors and also performs affiliation parsing. Extraction and parsing are done primarily by machine learning-based algorithms, which makes the system more adaptable to new layouts and styles. The web service, as well as the source code, are publicly available. We compare our solution to other tools with respect to author and affiliation extraction performance. (See Section 4.)
3 Author and Affiliation Extraction Workflow
The affiliation extraction algorithm accepts a scientific publication in PDF format as input and extracts authors and affiliations in structured form. The following is an example fragment of the algorithm's output in NLM JATS format:<contrib-group>
<contrib contrib-type="author">
<string-name>Antje Theurer</string-name>
<xref ref-type="aff" rid="1">1</xref>
<xref ref-type="aff" rid="2">2</xref>
</contrib>
<aff id="1">
<label>1</label>
<institution>Department of Internal Medicine, St. Katharinen Hospital</institution>,
<addr-line>Seckbacher Landstrasse 65, 60389, Frankfurt</addr-line>,
<country country="DE">Germany</country>
</aff>
<aff id="2">
<label>2</label>
<institution>Medical Department, Kamuzu Central Hospital</institution>,
<addr-line>Lilongwe</addr-line>
<country country="MW">Malawi</country>
</aff>
</contrib-group>
The extraction work flow is composed of the following steps, as shown in Figure 1:
- First, basic geometric structure is extracted from a PDF file. The structure stores the entire text content of the input document and the geometric features related to the way the text is displayed in the PDF file. More precisely, the structure is composed of pages, zones, lines, words and characters, along with their coordinates, dimensions and reading order. The structure can be serialized using TrueViz format [7].
- Next, we assign functional labels to all zones during a content classification step. Some examples of labels include: title, body, references, author, affiliation, correspondence.
- Zones containing authors and affiliations are split into individual elements and relations between them are derived.
- Finally, raw affiliation strings are parsed in order to detect their institution, address and country.
Table 1 shows the decomposition of the extraction work flow into individual steps and provides basic information about tools and algorithms used for each step.
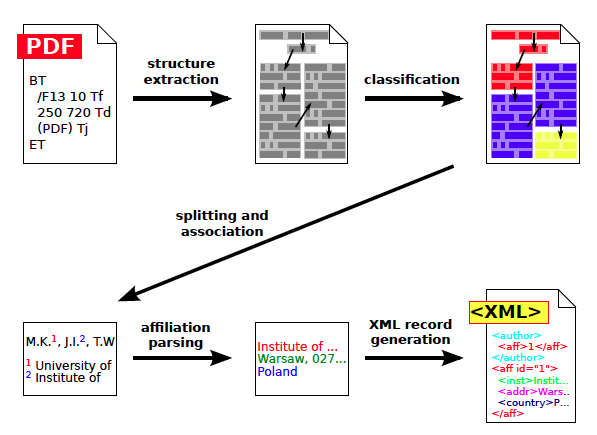
Figure 1: Overview of the affiliation extraction algorithm. At the beginning the basic structure is extracted from the PDF file, then document's fragments are classified. Next, detected fragments containing authors and affiliations are split and matched together. Finally, raw affiliation strings are parsed and the resulting XML record is formed. |
| Step | Goal | Implementation |
| Character extraction | Extracting individual characters along with their page coordinates and dimensions from the input PDF file. | iText library |
| Page segmentation | Constructing the document's geometric hierarchical structure containing (from the top level) pages, zones, lines, words and characters, along with their page coordinates and dimensions. | Enhanced Docstrum |
| Reading order resolving | Determining the reading order for all structure elements. | Bottom-up heuristics-based |
| Initial zone classification | Classifying the document's zones into four main categories: metadata, body, references and other. | Support Vector Machines |
| Metadata zone classification | Classifying the document's zones into specific metadata classes. | Support Vector Machines |
| Author and affiliation extraction | Extracting lists of authors and affiliations from labelled zones and associating authors with affiliations. | Heuristics-based |
| Affiliation parsing | Extracting organization name, address and country from affiliation strings. | Conditional Random Fields |
Table 1: The decomposition of the extraction work flow into individual steps.
3.1 Document structure extraction
Structure extraction is the initial phase of the entire work flow. Its goal is to create a hierarchical structure of the document preserving the entire text content of the input document and also features related to the way the text is displayed in the PDF file. More information about the implementation details can be found in [14].
Basic structure extraction is composed of three steps:
- Character extraction — extracting individual characters from the input PDF document.
- Page segmentation — joining individual characters into words, lines and zones.
- Reading order determination — calculating the reading order for all structure levels.
The purpose of character extraction is to extract individual characters from the PDF stream along with their positions on the page and dimensions. The implementation is based on the open-source iText library. We use iText to iterate over a PDF's text-showing operators. During the iteration we extract text strings along with their size and position on the page. Next, extracted strings are split into individual characters and their individual widths and positions are calculated. The result is an initial flat structure of the document, which consists only of pages and characters. The widths and heights computed for individual characters are approximate and can differ slightly from the exact values depending on the font, style and characters used. Fortunately, those approximate values are sufficient for further steps.
The goal of page segmentation is to create a geometric hierarchical structure storing the document's content. As a result the document is represented by a list of pages, each page contains a set of zones, each zone contains a set of text lines, each line contains a set of words, and finally each word contains a set of individual characters. Each object in the structure has its content, position and dimensions.
Page segmentation is implemented with the use of a bottom-up Docstrum algorithm [11]:
- The algorithm is based to a great extent on the analysis of the nearest-neighbor pairs of individual characters. In the first step, five nearest components for every character are identified (red lines in Figure 2).
- In order to calculate the text orientation (the skew angle) we analyze the histogram of the angles between the elements of all nearest-neighbor pairs. The peak value is assumed to be the angle of the text.
- Next, within-line spacing is estimated by detecting the peak of the histogram of distances between the nearest neighbors. For this histogram we use only those pairs in which the angle between components is similar to the estimated text orientation angle (blue lines in Figure 2).
- Similarly, between-line spacing is also estimated with the use of a histogram of the distances between the nearest-neighbor pairs. In this case we include only those pairs that are placed approximately in the line perpendicular to the text line orientation (green lines in Figure 2).
- Next, line segments are found by performing a transitive closure on within-line nearest-neighbour pairs. To prevent joining line segments belonging to different columns, the components are connected only if the distance between them is sufficiently small.
- The zones are then constructed by grouping the line segments on the basis of heuristics related to spatial and geometric characteristics: parallelness, distance and overlap.
- The segments belonging to the same zone and placed in one line horizontally are merged into final text lines.
- Finally, we divide the content of each text line into words based on within-line spacing.
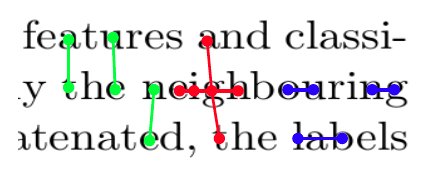
Figure 2: An example fragment of a text zone in a scientific article. The figure shows five nearest neighbours of a given character (red lines), neighbours placed in the same line used to determine inline spacing (blue lines), and neighbours placed approximately in the line perpendicular to the text line orientation used to determine between-line spacing (green lines). |
A PDF file contains, by design, a stream of strings that undergoes an extraction and segmentation process. As a result we obtain pages containing characters grouped into zones, lines and words, all of which form what could be described as an "unsorted bag of items". The aim of setting the reading order is to determine the right sequence in which all the structure elements should be read. An example document page with a reading order of the zones is shown in Figure 3.

Figure 3: An example page from a scientific publication. The image shows the zones and their reading order. |
A reading order resolving algorithm is based on a bottom-up strategy: first, characters are sorted within words and words within lines horizontally, then lines are sorted vertically within zones, and finally we sort zones. The fundamental principle for sorting zones was taken from PdfMiner. We make use of an observation that the natural reading order in most modern languages descends from top to bottom, if successive zones are aligned vertically, otherwise it traverses from left to right. This observation is reflected in the distances counted for all zone pairs: the distance is calculated using the angle of the slope of the vector connecting zones. As a result zones aligned vertically are in general closer than those aligned horizontally. Then using an algorithm similar to hierarchical clustering methods we build a binary tree by repeatedly joining the closest zones and groups of zones. After that, for every node its children are swapped, if needed. Finally, an in order tree traversal gives the desired zones order.
3.2 Content classification
The goal of content classification is to label each zone with a functional class. The classification is done in two stages: initial classification assigns general categories (metadata, references, body, other), while the goal of metadata classification is to classify all metadata zones into specific metadata classes (abstract, bib info, type, title, affiliation, author, correspondence, dates, editor, keywords).
Both classifiers use Support Vector Machines and their implementation is based on the LibSVM library [1]. The classifiers differ in SVM parameters, but in both cases the best parameters were found by performing a grid-search using a set of 100 documents from PubMed Central Open Access Subset and maximizing mean F-score obtained during a 5-fold cross validation.
In order to perform zone classification, each zone is transformed into a vector of feature values, which are to a great extent the same for both classifiers. The initial and metadata classifiers use 83 and 62 features, respectively. The features capture various aspects of the content and surroundings of the zones and can be divided into the following categories:
- geometric — based on geometric attributes, some examples include: zone's height and width, height to width ratio, zone's horizontal and vertical position, the distance to the nearest zone, empty space below and above the zone, mean line height, whether the zone is placed at the top, bottom, left or right side of the page;
- lexical — based upon keywords characteristic of different parts of a narration, such as: affiliations, acknowledgments, abstract, keywords, dates, references, article type; these features typically check whether the text of the zone contains any of the characteristic keywords;
- sequential — based on sequence-related information; some examples include: the label of the previous zone (according to the reading order), the presence of the same text blocks on the surrounding pages, whether the zone is placed in the first/last page of the document;
- formatting — related to text formatting in the zone; examples include: font size in the current and adjacent zones, the amount of blank space inside zones, mean indentation of text lines in the zone;
- heuristics — based on heuristics of various kinds, such as the count and percentage of lines, words, uppercase words, characters, letters, upper/lowercase letters, digits, whitespaces, punctuation, brackets, commas, dots, etc.; also whether each line starts with enumeration-like tokens or whether the zone contains only digits.
3.3 Author and affiliation metadata extraction
As a result of classifying the document's fragments, we usually obtain a few regions labelled as author or affiliation. In this step we extract individual author names and affiliations and determine relations between them.
In general the implementation is based on heuristics and regular expressions, but the details depend on the article's layout. There are two main styles used in different layouts: (1) the names of all authors are placed in one zone in the form of a list, and similarly affiliations are listed together below the authors' list, at the bottom of the first page or at the end of the document, before the bibliography section (an example is shown in Figure 4), and (2) each author name is placed in a separate zone along with its affiliation and email address (an example is shown in Figure 5).

Figure 4: An example fragment of a page from a scientific publication with author names and affiliations zones. In this case the relations author-affiliation (coded with colors) can be determined with the use of separators and upper indexes. |
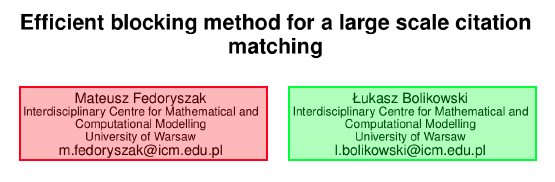
Figure 5: An example fragment of a page from a scientific publication with authors and affiliations zones. In this case the relations author-affiliation can be determined using the distance and other geometric features of the text. |
At the beginning, the algorithm recognizes the type of layout of a given document. If the document contains two zones labelled as affiliation placed horizontally, it is treated as type (2), otherwise as type (1).
In the case of a layout of the first type (Figure 4), at the beginning authors' lists are split using a predefined lists of separators. Then we detect affiliation indexes based on predefined lists of symbols and also geometric features. Detected indexes are then used to split affiliation lists and assign affiliations to authors.
In the case of a layout of the second type (Figure 5), each author is already assigned to its affiliation by being placed in the same zone. It is therefore enough to split the content of such a zone into author name, affiliation and email address. We assume the first line in the zone is the author name, email is detected based on regular expressions, and the rest is treated as the affiliation string. In the future we plan to implement this step using a supervised token or line classifier.
3.4 Affiliation parsing
The goal of affiliation parsing is to recognize affiliation string fragments related to institution, address and country. Additionally, country names are decorated with their ISO codes. Figure 6 shows an example of a parsed affiliation string.

Figure 6: An example of a parsed affiliation string. Colors mark fragments related to institution, address and country.
Affiliation parser uses Conditional Random Fields classifier and is built on top of GRMM and MALLET packages [10]. The first affiliation string is tokenized, then each token is classified as institution, address, country or other, and finally neighbouring tokens with the same label are concatenated. The token classifier uses the following features:
- the classified word itself,
- whether the token is a number,
- whether the token is an all uppercase/lowercase word,
- whether the token is a lowercase word that starts with an uppercase letter,
- whether the token is contained in dictionaries of countries or words commonly appearing in institutions or addresses.
The class of a given token depends not only on its features and classes of surrounding tokens, but also on their features. To reflect this in the classifier, the token's feature vector contains not only features of the token itself, but also features of two preceding and two following tokens.
4 Evaluation
We performed the evaluation of the key steps of the algorithm and the entire extraction process as well. The ground truth data used for the evaluation is based on the resources of PubMed Central Open Access Subset.
4.1 Datasets preparation
Three datasets were used during the evaluation process. A subset of PMC was used directly to evaluate affiliation extraction work flow. Additionally, PMC served as a base for constructing a GROTOAP2 dataset, which was used for training and evaluating zone classifiers. A separate affiliation dataset was used to train and evaluate the affiliation parser.
PubMed Central Open Access Subset contains life sciences publications in PDF format accompanied by corresponding metadata in the form of XML NLM files. NLM files contain a rich set of documents' metadata, structured full text and also documents' bibliography. A subset of 1,943 documents from PMC was directly used to evaluate the entire affiliation extraction process.
Unfortunately, NLM files contain only the annotated text of documents and do not preserve geometric features related to the way the text is displayed in PDF files. As a result, PMC could not be used directly for training and evaluation of zone classifiers. For these tasks we built GROTOAP2 [13] — a large and diverse dataset containing 13,210 documents from 208 different publishers. The main part of the dataset are semi-automatically created ground truth files in TrueViz format. They contain the entire text content of publications in hierarchical form composed of pages, zones, lines, words and characters, their reading order and zone labels.
Affiliation dataset used for parser evaluation contains 8,267 parsed affiliations from PMC documents. Since parsed affiliations in NLM files do not always contain the entire raw affiliation string as it was given in the article, we built the dataset automatically by matching labelled metadata from NLM documents against affiliation strings extracted from PDFs by CERMINE. The dataset is publicly available here.
4.2 Classification evaluation
Both zone classifiers were evaluated by a 5-fold cross validation using a set of zones from 2,551 documents from the GROTOAP2 dataset. The set contains 355,779 zones, 68,557 of which are metadata zones. Tables 2 and 3 show the results for labels of interest: metadata and affiliation.
| metadata | other labels | precision | recall | |
| metadata | 66,372 | 2,185 | 96.8% | 97.0% |
| other labels | 2,052 | 285,170 | - | - |
Table 2: The results of initial classification for 5-fold cross validation. Rows and columns represent the desired and obtained classification result, respectively. |
| affiliation | other labels | precision | recall | |
| affiliation | 3,496 | 185 | 95.0% | 95.3% |
| other labels | 173 | 64,703 | - | - |
Table 3: The results of metadata classification for 5-fold cross validation. Rows and columns represent the desired and obtained classification result, respectively. |
4.3 Affiliation parsing evaluation
Affiliation parser was evaluated by a 5-fold cross validation with the use of 8,267 affiliations from PMC. The confusion matrix for token classification is shown in Table 4.
| address | country | institution | precision | recall | |
| address | 44,481 | 12 | 1,225 | 96.8% | 97.3% |
| country | 50 | 8,108 | 8 | 99.6% | 99.3% |
| institution | 1,434 | 18 | 92,457 | 98.7% | 98.5% |
Table 4: Confusion matrix for affiliation token classification for 5-fold cross validation. Rows and columns represent the desired and obtained classification result, respectively. |
In addition to evaluating individual token classification, we also checked for how many affiliations the entire fragments (institution, address or country) were labelled correctly. The following results were obtained:
- institution was correctly recognized in 92.4% of cases,
- address was correctly recognized in 92.2% of cases,
- country was correctly recognized in 99.5% of cases,
- 92.1% of affiliations were entirely correctly parsed.
4.4 Affiliation extraction workflow evaluation
A subset of 1,943 documents from PMC was used to compare the performance of various extraction systems. We evaluated four tasks:
- extracting author strings from a given article,
- extracting affiliation strings from a given article,
- determining author-affiliation relations in a given article,
- determining author-affiliation relations, provided that authors and affiliations were extracted flawlessly.
During the evaluation we used the default version of CERMINE, in which zone classifiers are trained on a subset of the GROTOAP2 dataset and the affiliation parser is trained on our affiliation dataset.
Apart from CERMINE, the following systems were evaluated: GROBID, ParsCit and PDFX. We used the default versions of the tools, without retraining them or modifying them in any way. PDFX was executed through its web service.
The tools processed articles in PDF format. The only exception was ParsCit, which analyses only the text content of a document, and therefore, in this case, PDF files were first transformed to text using pdftotext tool. The results were obtained by comparing the output provided by the tools with annotated data from NLM files.
For every document and every task we compared the lists of extracted and ground truth objects. Author and affiliation strings were compared using cosine distance with a threshold. In the case of author-affiliation relations, a relation was marked as correct if both elements matched. This resulted in individual precision and recall for every document. The overall precision and recall values were computed as mean values over all documents from the dataset.
The evaluation results are shown in Figure 7. In both author and affiliation extraction tasks CERMINE achieved the best F-score (89.2% and 84.3%, respectively). In the case of determining author-affiliation relations we compared our solution only to GROBID, as other tools do not extract this information. In both tasks CERMINE achieved better results (F1 63.1% and 77.4% in tasks 3 and 4, respectively).
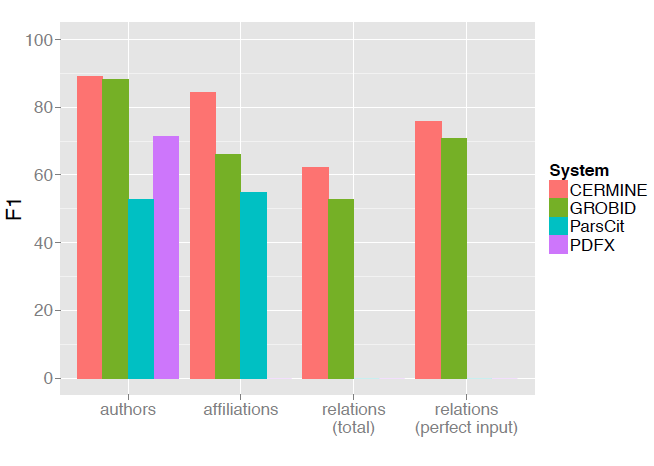
Figure 7: The results of metadata extraction comparison. The plot shows F1 scores obtained by various extraction tools in each of the four tasks: 1) extracting author strings, 2) extracting affiliation strings, 3) determining author-affiliation relations, and 4) determining author-affiliation relations when authors and affiliations were extracted without errors. Missing values appear in the case of tools which do not extract a given metadata type. |
5 System Usage
CERMINE's web service is available online and can be accessed here. The source code is available on GitHub here. The system also provides RESTful services that allow for executing the extraction and parsing tasks by machines, using tools like cURL.
Metadata can be extracted from a scientific article in PDF format using the following command:
$ curl -X POST --data-binary @article.pdf \
--header "Content-Type: application/binary" -v \
http://cermine.ceon.pl/extract.do
Affiliation string can be parsed using the following command:
$ curl -X POST \
--data "affiliation=the text of the affiliation" \
http://cermine.ceon.pl/parse.do
The time needed to extract metadata from a document depends mainly on its number of pages. Figure 8 shows the processing time as a function of the number of pages for 1,238 random documents. The average processing time for this subset was 9.4 seconds.
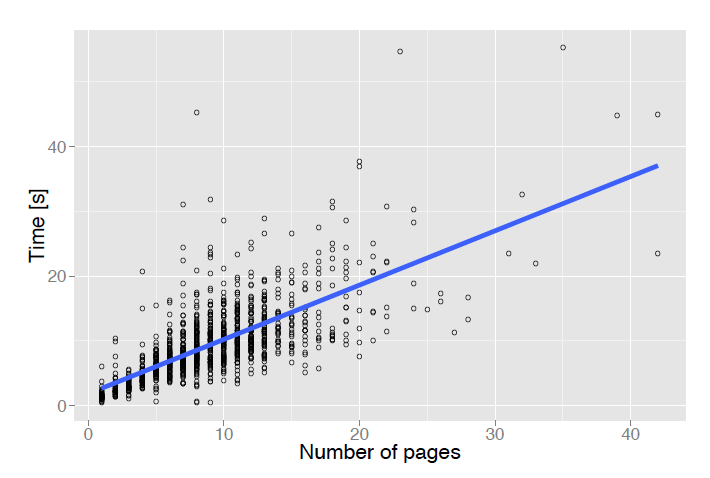
Figure 8: Metadata extraction time as a function of a document's number of pages.
Page segmentation and initial zone classification are the most time-consuming steps. By default CERMINE processes the entire input document in order to extract its full text. However, if the client application is interested in document metadata only (for example document title, authors and affiliations), it is sufficient to analyse only the first and last pages of the document as the metadata is rarely present in the middle part. In such cases we recommend restricting the analysis to a fixed number of pages, and as a result even large documents can be processed in a reasonable time.
6 Conclusion
This article described an affiliation extraction method for extracting authors and affiliations from scientific publications in PDF format, associating authors with affiliations, and parsing affiliation strings in order to extract institution, address and country information. The algorithm used is a part of the open source CERMINE system, a tool for extracting structured metadata and content from born-digital scientific literature. According to our evaluation of the key steps of the algortihm, as well as the entire extraction process, we found that our solution performs the best in all of the evaluated tasks. The method is effective and reliable and can be used whenever we do not have access to valuable author and affiliation metadata. Our future plans include comparing a heuristic-based approach with machine learning for zone content splitting and determining author-affiliation relations.
Acknowledgements
This work has been partially supported by the European Commission as part of the FP7 project OpenAIREplus (grant no. 283595).
References
[1] C. Chang and C. Lin. LIBSVM: A library for support vector machines. ACM TIST, 2(3):27, 2011. http://doi.org/10.1145/1961189.1961199
[2] A. Constantin, S. Pettifer, and A. Voronkov. PDFX: fully-automated pdf-to-xml conversion of scientific literature. In ACM Symposium on Document Engineering, pages 177-180, 2013. http://doi.org/10.1145/2494266.2494271
[3] I. G. Councill, C. L. Giles, and M. Kan. Parscit: an open-source CRF reference string parsing package. In Proceedings of the International Conference on Language Resources and Evaluation, LREC 2008, 26 May - 1 June 2008, Marrakech, Morocco, 2008.
[4] H. H. N. Do, M. K. Chandrasekaran, P. S. Cho, and M. Y. Kan. Extracting and matching authors and affliations in scholarly documents. In Proceedings of the 13th ACM/IEEE-CS Joint Conference on Digital Libraries, JCDL '13, 2013. http://doi.org/10.1145/2467696.2467703
[5] H. Han, C. L. Giles, E. Manavoglu, H. Zha, Z. Zhang, and E. A. Fox. Automatic document metadata extraction using support vector machines. In ACM/IEEE 2003 Joint Conference on Digital Libraries (JCDL 2003), 27-31 May 2003, Houston, Texas, USA, Proceedings, pages 37-48, 2003. http://doi.org/10.1109/JCDL.2003.1204842
[6] S. Jonnalagadda and P. Topham. NEMO: extraction and normalization of organization names from pubmed affliation strings. CoRR, abs/1107.5743, 2011.
[7] Chang Ha Lee, Tapas Kanungo. The architecture of TrueViz: a groundTRUth/metadata editing and VIsualiZing ToolKit. Pattern Recognition 36(3): 811-825 (2003) http://doi.org/10.1016/S0031-3203(02)00101-2
[8] P. Lopez. GROBID: combining automatic bibliographic data recognition and term extraction for scholarship publications. In Research and Advanced Technology for Digital Libraries, 13th European Conference, pages 473-474, 2009. http://doi.org/10.1007/978-3-642-04346-8_62
[9] M. Luong, T. D. Nguyen, and M. Kan. Logical structure recovery in scholarly articles with rich document features. IJDLS, 1(4):1-23, 2010. http://doi.org/10.4018/jdls.2010100101
[10] A. K. McCallum. MALLET: A Machine Learning for Language Toolkit. 2002.
[11] L. O'Gorman. The document spectrum for page layout analysis. IEEE Trans. Pattern Anal. Mach. Intell., 15(11):1162-1173, 1993. http://doi.org/10.1109/34.244677
[12] D. Tkaczyk et al. Cermine: Cermine 1.6, May 2015. http://doi.org/10.5281/zenodo.17594
[13] D. Tkaczyk, P. Szostek, and L. Bolikowski. GROTOAP2 - the methodology of creating a large ground truth dataset of scientific articles. D-Lib Magazine, 2014. http://doi.org/10.1045/november14-tkaczyk
[14] D. Tkaczyk, P. Szostek, M. Fedoryszak, P. J. Dendek, and L. Bolikowski. Cermine: automatic extraction of structured metadata from scientific literature. International Journal on Document Analysis and Recognition (IJDAR), pages 1-19, 2015. http://doi.org/10.1007/s10032-015-0249-8
About the Authors
 |
Dominika Tkaczyk is a researcher at the Interdisciplinary Centre for Mathematical and Computational Modelling at University of Warsaw (ICM UW). She received her MSc in Computer Science from University of Warsaw. Her current research interests focus on extensive analysis of scientific literature. She is the lead developer of CERMINE — a machine learning-based Java library for extracting metadata and content from scholarly publications. She has also contributed to the design and development of a big data knowledge discovery service for OpenAIREplus project. Previously she was involved in developing YADDA2 system — a scalable open software platform for digital library applications. |
 |
Bartosz Tarnawski studies Mathematics and Computer Science at University of Warsaw. He is interested in discrete mathematics, especially graph theory. During his internship at Interdisciplinary Centre for Mathematical and Computational Modelling (ICM UW) he joined the CERMINE project and participated in the development of affiliation parsing functionality. |
 |
Łukasz Bolikowski is an Assistant Professor at the Interdisciplinary Centre for Mathematical and Computational Modelling at University of Warsaw (ICM UW). He defended his PhD thesis on semantic network analysis at Systems Research Institute of Polish Academy of Sciences. He is a leader of a research group focusing on scalable knowledge discovery in scholarly publications. He has contributed to a number of European projects, including DRIVER II, EuDML, OpenAIREplus. Earlier at ICM UW he specialized in construction of mathematical models and their optimization on High Performance Computing architectures.. |
|
|
|
| P R I N T E R - F R I E N D L Y F O R M A T | Return to Article |