|
|
|
| P R I N T E R - F R I E N D L Y F O R M A T | Return to Article |
D-Lib Magazine
March/April 2016
Volume 22, Number 3/4
Harvesting and Repurposing Metadata from Web of Science to an Institutional Repository Using Web Services
Yuan Li
Princeton University
[email protected]
DOI: 10.1045/march2016-li
Abstract
This article describes the workflow used to ingest faculty publication citations into a Digital Commons platform Institutional Repository, by repurposing metadata harvested from the Web of Science using web services. This approach, along with a workflow, were developed to improve the populating process for the IR. Improving upon the previous manual process, this approach automates most of the collecting and ingesting process and greatly improves ingesting efficiency. A discussion of the benefits and limitations of the new approach is also provided.
1 Introduction
One of the core functions of an institutional repository (IR) is to collect, preserve, and promote faculty research output. Most importantly, an IR provides access to alternative formats (pre-prints or post-prints) of research content for people who can't afford subscriptions or have a limited capacity to access the research output. The manual process of collecting and ingesting metadata into an IR is time-consuming, especially for an institution with a large number of faculty but very thin IR staffing.
In 2007, the University of Massachusetts Amherst's W.E.B. Du Bois Library established its IR, ScholarWorks@UMassAmherst, using Digital Commons (an IR hosting platform) from Berkeley Electronic Press (bepress), for archiving a broad range of scholarly output including faculty research, theses and dissertations, working papers, technical reports, books and more. The process that was developed to collect faculty scholarship shown in Figure 1 below, is similar to that developed by many other institutions.
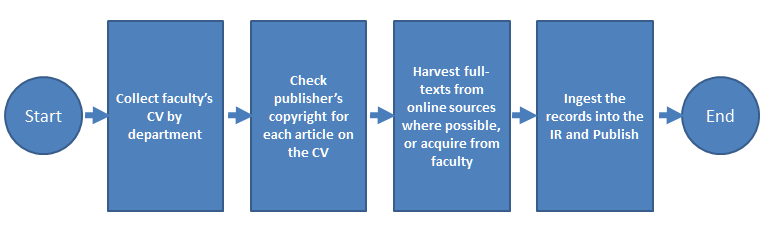
Figure 1: IR Content Process Flow Chart
First, the faculty's CV is collected. Then the publisher's copyright for each citation on the CV is checked. Subscription databases and open access subject repositories are searched in order to find a copy of the article to archive. If a copy is not available, a subject liaison requests one from the faculty member. After obtaining the citation and the full-text, the records are manually entered into the IR. The entire process is primarily manual, which presents staff workload challenges for libraries when populating an IR. The UMass Amherst library recognized that automation of collection and ingest was key to improving efficiency.
While working as a member of the Scholarly Communication Office at UMass Amherst, the author developed a new approach to collecting and ingesting the faculty publication citations, using open source tools. Although the approach only applies to the last step in the ingest process (see Figure 2 below), the efficiency of the entire process was significantly improved.
This article is based on a presentation made by the author at the Library Information Technology Association (LITA) program held during 2015 ALA Annual Conference. Although the work was done in 2010, the approach remains relevant and useful in current practice. Although the approach and the workflow described were developed for IRs using the Digital Commons platform, it can be easily adapted for repositories created on other platforms such as DSpace or Eprint.1
2 Process for Harvesting and Ingest Citations
Before beginning the harvesting and ingest process, the first step is to connect web services from local servers to the Web of Science (WOS) server, which is the core technology used by ScholarWorks@UMassAmherst for automating the harvesting and ingesting process. In order to use the web services from WOS, an institution must have WOS membership or a subscription. The web services setup only needs to be done once. After the setup is completed, the ingesting process or workflow is fairly straightforward, including building connections, harvesting (using web services), transforming metadata harvested from Web of Science, and batch ingesting into the repository. Figure 2 illustrates the workflow.
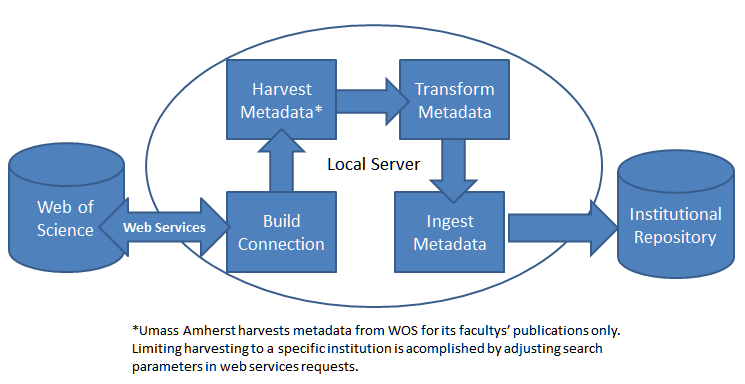
Figure 2: Harvesting and Ingesting Workflow Chart
2.1 Tools
The process described below uses SoapUI, an open source web services tool, and Notepad++, an open source XML editing tool, both of which can be downloaded and installed for free.
2.2 Set up Web of Science (WOS) Web Services
The Web of Science's documentation describes its Web Service as "a program that allows two computer systems to interact over a network. The Web of Science Web Services allows a customer's system to request information from Web of Science Core Collection and returns a limited set of data to the requesting system."2
Web Services was used to batch extract data from WOS in a web services tool, SoapUI. Web of Science makes available Web of Knowledge Web Services Lite V.3.03, a document that explains what web services they offer, the detailed operations, and the data (metadata fields) retrieved from Web of Science.
The web services setup includes software configuration and projects setup. A project is an operation which has to be performed in order to build a successful connection between the Web of Science server and the local server, to enable communication between the servers. Two projects need to be set up, one for authentication and another for search. As stated earlier, before starting to harvest any data from the WOS server, we needed to first build the connection between the local server and the WOS server. The authentication project is the operation to make the connection happen. The search project is the operation to harvest data. Search enables telling the WOS server what data the WOS server should return. The search code is written in Soap and XML. (The code template is provided in the Appendices). Two endpoint addresses, both provided by WOS, are required for the operations to be successful:4
- Authenticate service endpoint address: http://search.isiknowledge.com/esti/wokmws/ws/WOKMWSAuthenticate?wsdl
- Search service endpoint address: http://search.isiknowledge.com/esti/wokmws/ws/WokSearchLite?wsdl
Once these two projects are completed, the rest of the workflow can be set up without difficulty. Detailed instructions for Web Services setup is included in the Appendices.
2.3 Build Connection and Harvest Metadata from Web of Science (WOS) by Individual Faculty Member
The workflow starts by building a connection between servers. Creation of a "session identifier" indicates the connection is successful.
- Authenticate operation
Open SoapUI, click "authenticate project", then click "authenticate Request." Two windows (also called working space) will appear. The one on the left is for "Request". This is where the authentication request is written in XML. The window on the right is for the "Response". This is where data from the server will be retrieved after the request is sent. For the authentication operation, the following code is used to request authentication from the Web of Science server. Once the request is sent and successfully authenticated, a "session identifier" will be returned in the response window on the left. This "session identifier" is the key for continuing to the next step, the search.
<soapenv:Header/>
<soapenv:Body>
<ns2:authenticate xmlns:ns2="http://auth.cxf.wokmws.thomsonreuters.com"/>
</soapenv:Body>
</soapenv:Envelope>
- Search operation (harvest data)
Once a successful connection is made, the harvesting process can begin searching by individual faculty member. The following code is used to query the data from the server. Although most of the code is default, parameters can be adjusted to customise the search. For example, the search time period can be adjusted (<timeSpan></timeSpan>); the author can be changed by changing the value of "AU" between tags <userQuery></userquesry>; the number of results requested adn returned can be changed (between tags <count></count>) and in what order (Ascending or Descending between tags <sort></sort>). The detailed workflow included in the Appendices explains how to perform the search operation step by step.
<soap:Header/>
<soap:Body>
<ns2:search xmlns:ns2="http://woksearchlite.cxf.wokmws.thomsonreuters.com">
<queryParameters>
<databaseID>WOS</databaseID>
<editions>
<collection>WOS</collection>
<edition>SCI</edition>
</editions>
<editions>
<collection>WOS</collection>
<edition>ISTP</edition>
</editions>
<queryLanguage>en</queryLanguage>
<timeSpan>
<begin>1980-07-01</begin>
<end>2010-06-30</end>
</timeSpan>
<userQuery>OG=Univ Massachusetts AND CI=Amherst AND AU=Malone, M*></userQuery>
</queryParameters>
<retrieveParameters>
<count>100</count>
<fields>
<name>Date</name>
<sort>D</sort>
</fields>
<firstRecord>1</firstRecord>
</retrieveParameters>
</ns2:search>
</soap:Body>
</soap:Envelope>
2.4 Transform Web Of Science schema to Digital Commons schema using in-house developed XSL Stylesheet
Table 1 below is used to map the metadata from the Web of Science schema to a Digital Commons schema. For elements from the Web of Science schema that are not represented explicitly in the Digital Commons schema, we use a custom field element. The XSL stylesheet is used to transform the metadata XML files from one schema to another using Notepad++. The in-house developed XSL Stylesheet, Notepad++ installation instruction, and the WOS to DC XML Schema Converting instructions are shared in the Appendices. The metadata harvested from Web of Science is usually in XML by individual faculty member. When transforming the XML metadata files, the transformation can be done by individual faculty member or it can be done in a batch depending on how your repository communities/collections are organized. For the latter (transforming files in a batch), the multiple XML files need to be combined into one XML file. The method for combining multiple XML files into one was introduced in Averkamp and Lee's article in 20095.
Table 1: Mapping Metadata from Web of Science Schema to Digital Commons Schema
| Web of Science (source) | Digital Commons (output) | Notes |
| return | documents | root |
| records | document | item |
| title/@label="Title"/values | title | Article title |
| authors/@label="Authors"/values | authors/author/lname authors/author/fname |
authors |
| Keywords/@label="Keywords"/values | keywords/keyword | Keywords |
| Source/@label="Issue"/values | Fields/field/@name="issue"/value | Issue |
| Source/@label="volume"/values | Fields/field/@name="volume"/value | Volume |
| Source/@label="Pages"/values | Fields/field/@name="pages"/value | Page |
| Source/@label="Pbulished.BiblioDate"/values Source/@label="Published.BiblioYear"/values |
Fields/field/@name="publication_date_date_format"/value | Publication dates |
| Source/@label="SourceTitle"/values | Fields/field/@name="journal_title"/value | Journal title |
2.5 Batch upload citations in XML to repository
Finally, the XML batch upload function6 in Digital Commons is used to ingest the metadata of faculty articles into the appropriate department community.
3 Benefits & Limitations
The main benefit of using web services to harvest the metadata is that it greatly improves the efficiency of the records ingestion workflow. Using the manual workflow one staff member or student worker can ingest up to 50 records into the repository, while in the same period of time, the number of records ingested goes up to 1,000 when the person uses the new workflow. The manual ingestion workflow had been a bottleneck affecting the entire repository production process. After deploying the web services technology, the process became much more efficient.
However, there are some limitations to the approach. The first is that the approach only automates the last step in the process of populating an IR. The other steps, such as checking publisher's copyright and full-text acquisition, are still primarily manual and do not benefit from this new approach.
The second limitation involves the web services setup. Although detailed setup instructions are provided, some upfront technical work is still required. Someone without a solid technical background would need help from IT support personnel as well as technical support from Web of Science.
Thirdly, the training can be challenging for staff or the student workers at the beginning. The detailed operational steps are provided in the Appendices. A few steps that involve manual correction in the code might be intimidating to someone who doesn't feel comfortable working with lines of code. However, having gone through the workflow once or twice, it should become routine.
Fourthly, the workflow is limited in the scope of its coverage. The workflow was developed for only one database, Web of Science. Web of Science is a big database covering many disciplines with a considerable span of years, but it doesn't cover everything. For records not covered by Web of Science, extra work is needed to get them into the repository.
Lastly, this approach only harvests publications' metadata, not the full-text. Full-text acquisition and uploading is a separate step with a different workflow. For Digital Commons users, batch uploading full-text function is available7.
4 Conclusion
The workflow described in this article has greatly improved the efficiency of repository ingestion. Although there are limitations, it has helped us improve the repository production process as a whole. With the passage of time, however, Web of Science may change the way they deliver their metadata by using technology other than web services. If this occurs, the workflow will need to be revisited and changed accordingly. At the same time, a few other harvesting (metadata or full-text) services have been developed for free or for a fee, such as ARL SHARE notification services (in the pilot phase) and services from Symplectic Element. The workflow described in this article may at some point in the future be replaced by one of those harvesting services.
Acknowledgements
During the preparation of the paper, I received a lot of support from our colleagues. Here I would like to thank Neil Nero, Behavioral Science Librarian from Lewis Library, Princeton University, Willow Dressel, Engineering Librarian from Engineering Library, Princeton University, and Charlotte Roh, Digital Repository Resident from W.E. B. Du Bios Library, UMass Amherst for their editorial suggestion.
Notes
| 1 | The detailed workflow plus web services setup instruction, XSLT stylesheet, and records examples (in XML) are shared as Appendices for others to use and adapt. |
| 2 | Web of Science, Web Services FAQ. |
| 3 | Thomson Reuters. (2012). Web of Knowledge Web Services Lite V. 3.0. |
| 4 | The authenticate and search service endpoint address is only available to WOS subscribers. An organization's WOS representative(s) can assist and provnamee further technical support. |
| 5 | Averkamp, S. and Lee, J. (2009). Repurposing ProQuest Metadata for Batch Ingesting ETDs into an Institutional Repository. Code4Lib Journal, 7. |
| 6 | Batch uploading records in xml is supported by Digital Commons. Contact DC client services for instructions. |
| 7 | Digital Commons. (2012). Batch Upload, Export and Revise with Excel: How to Manage Bulk Records in a Digital Commons. |
Appendices
Appendices include a detailed workflow, web services setup instruction, an XSLT stylesheet, and record examples (in XML).
| Appendix 1: | Web of Science to Digital Commons Semi-auto Harvesting Workflow |
| Appendix 2: | Instructions showing how ISI Web of Knowledge Web Services works with ScholarWorks |
| Appendix 3: | Notepad++ Installation Instructions |
| Appendix 4: | Instructions for Converting Web of Science (WOS) XML Schema to Digital Commons (DC) XML Schema |
| Appendix 5: | Digital Commons Record —Sample XML Files |
| Appendix 6: | WOS to DC Stylesheet |
| Appendix 7: | WOS Record XML File Sample |
About the Author
 |
Yuan Li is the Scholarly Communications Librarian at Princeton University, where she manages the Princeton University Library's efforts to support scholarly publication innovations and reforms, and supervises and coordinates activities related to the Princeton Open Access policy and the Princeton Institutional Repository. Prior to joining Princeton, she served as the Scholarly Communication Librarian at Syracuse University, the Digital Initiatives Librarian at University of Rhode Island, and the Digital Repository Librarian at University of Massachusetts Amherst. Yuan has a Master degree in Library and Information Science from the University of Rhode Island, a Master of Engineering degree in Applied Computer Science from the National Computer System Engineering Research Institute of China and a Bachelor of Engineering degree in Computer Science and Technology from Yanshan University (China). |
|
|
|
| P R I N T E R - F R I E N D L Y F O R M A T | Return to Article |